class: center, middle, inverse, title-slide # Joining data: <br> Messy 4 --- ## Messy (4) <br> **Messy (4)**: Values for a single observational unit are stored across multiple tables. <br> After data are normalized by splitting, we want to de-normalize again by *joining* datasets. The variables used to connect each pair of tables are the **keys**. --- ## Relational data It is not uncommon for a data analysis to involve many tables of data and you must combine them to answer the questions that you’re interested in. Collectively, multiple tables of data are called **relational data** because it is the relations, not just the individual datasets, that are important. To work with relational data you need **verbs** that work with pairs of tables. There are three families of verbs designed to work with relational data: 1. **Mutating joins**, which add new variables to one data frame from matching observations in another. 2. **Filtering joins**, which filter observations from one data frame based on whether or not they match an observation in the other table. 3. **Set operations**, which treat observations as if they were set elements. --- class: inverse, middle, center # Joining data from different tables --- ## Example: Lahman package Sean Lahman is a database journalist, who started databases of historical sports statistics, in particular, the Lahman database on baseball. ```r library(Lahman) LahmanData %>% head() ``` ``` ## file class nobs nvar title ## 1 AllstarFull data.frame 4993 8 AllstarFull table ## 2 Appearances data.frame 99466 21 Appearances table ## 3 AwardsManagers data.frame 171 6 AwardsManagers table ## 4 AwardsPlayers data.frame 6026 6 AwardsPlayers table ## 5 AwardsShareManagers data.frame 401 7 AwardsShareManagers table ## 6 AwardsSharePlayers data.frame 6705 7 AwardsSharePlayers table ``` --- ## Lahman database The Lahman database consists of 24 data frames that are linked by `playerID`. This is clean, but not very readable. The `Master` table includes names and other attributes for each player. *Joining* multiple tables helps us to bring together (pieces of) information from multiple sources. --- ## Example: Hall of Fame Who was inducted in the Hall of Fame in 2017? ```r HallOfFame %>% dplyr::filter(yearID == 2017) %>% head() ``` ``` ## playerID yearID votedBy ballots needed votes inducted category needed_note ## 1 bagweje01 2017 BBWAA 442 332 381 Y Player <NA> ## 2 raineti01 2017 BBWAA 442 332 380 Y Player <NA> ## 3 rodriiv01 2017 BBWAA 442 332 336 Y Player <NA> ## 4 hoffmtr01 2017 BBWAA 442 332 327 N Player <NA> ## 5 guerrvl01 2017 BBWAA 442 332 317 N Player <NA> ## 6 martied01 2017 BBWAA 442 332 259 N Player <NA> ``` --- class: yourturn # Your turn For this your turn use the `HallOfFame` and `Master` data from the `Lahman` package - Identify all players who were inducted in the Hall of Fame in 2017, by filtering the `Master` data for their player IDs. --- ## Joins: general idea 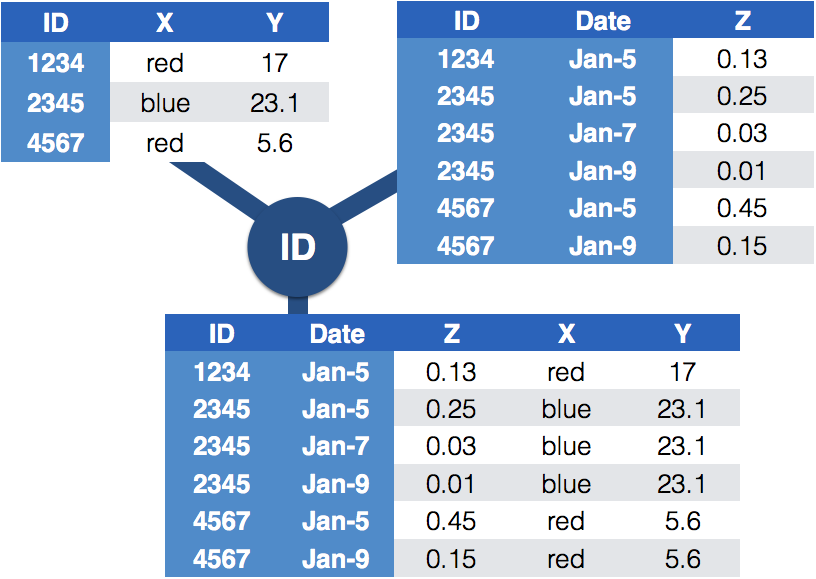 --- ## Joins: more specific idea - Data sets are joined along values of variables. - In `dplyr` there are several join functions: `left_join()`, `inner_join()`, `full_join()`, ... - The differences between join functions are only visible if values in one set do not all have values in the other set. --- ## Example data ```r df1 <- data.frame(id = 1:6, trt = rep(c("A", "B", "C"), rep=c(2,1,3)), value = c(5,3,7,1,2,3)) df1 ``` ``` ## id trt value ## 1 1 A 5 ## 2 2 B 3 ## 3 3 C 7 ## 4 4 A 1 ## 5 5 B 2 ## 6 6 C 3 ``` </br> ```r df2 <- data.frame(id=c(4,4,5,5,7,7), stress=rep(c(0,1), 3), bpm = c(65, 125, 74, 136, 48, 110)) df2 ``` ``` ## id stress bpm ## 1 4 0 65 ## 2 4 1 125 ## 3 5 0 74 ## 4 5 1 136 ## 5 7 0 48 ## 6 7 1 110 ``` --- ## Mutating joins A mutating join allows you to combine variables from two tables. We first match observations by their keys, then copy across variables from one table to the other. There are 4 mutating join functions: - `left_join(x, y, by = "key")`: keeps all observations in `x` - `right_join(x, y, by = "key")`: keeps all observations in `y` - `full_join(x, y, by = "key")`: keeps all observations in `x` and `y` - `inner_join(x, y, by = "key")`: keeps only observations that appear in both `x` and `y` --- ## `left_join(x, y, by = "key")` .pull-left-small[ ```r left_join(df1, df2, by="id") ``` ``` ## id trt value stress bpm ## 1 1 A 5 NA NA ## 2 2 B 3 NA NA ## 3 3 C 7 NA NA ## 4 4 A 1 0 65 ## 5 4 A 1 1 125 ## 6 5 B 2 0 74 ## 7 5 B 2 1 136 ## 8 6 C 3 NA NA ``` ] .pull-right-large[ - all elements in the *left* data set are kept - non-matches are filled in by `NA` - most commonly used; should be your default - use this when you look up additional data from another table since it preserves the original observations even when there isn’t a match - `right_join()` works symmetric ] --- ## `inner_join(x, y, by = "key")` .pull-left-small[ ```r inner_join(df1, df2, by = "id") ``` ``` ## id trt value stress bpm ## 1 4 A 1 0 65 ## 2 4 A 1 1 125 ## 3 5 B 2 0 74 ## 4 5 B 2 1 136 ``` ] .pull-right-large[ - ONLY matches pairs of observations whenever their keys are equal - output is a new data frame that contains the key, the x values, and the y values. - **most important property**: unmatched rows are NOT included in the result. - inner joins are usually not appropriate for use in analysis because it is too easy to lose observations. ] --- ## `full_join(x, y, by = "key")` .pull-left-small[ ```r full_join(df1, df2, by = "id") ``` ``` ## id trt value stress bpm ## 1 1 A 5 NA NA ## 2 2 B 3 NA NA ## 3 3 C 7 NA NA ## 4 4 A 1 0 65 ## 5 4 A 1 1 125 ## 6 5 B 2 0 74 ## 7 5 B 2 1 136 ## 8 6 C 3 NA NA ## 9 7 <NA> NA 0 48 ## 10 7 <NA> NA 1 110 ``` ] .pull-right-large[ - all ids are kept, missings are filled in with `NA` ] --- class: yourturn # Your turn For this exercise, use the data from the `Lahman` package. - Join (relevant pieces of) the `Master` data set and the `HallOfFame` data. - Find all Hall of Famers who were alive as of 2019. <br>(use the data resulted from the joining done in question 1) - How many attempts at being inducted to the HoF does Sammy Sosa have already? (use the data resulted from the joining done in question 1) --- ## Traps of joins - sometimes we unexpectedly cannot match values: missing values, different spelling, ... - join can be along multiple variables, e.g. `by = c("ID", "Date")` - joining variable(s) can have different names, e.g. `by = c("State" = "Name")` - always make sure to check dimensions of data before and after a join - check on missing values: - to get help with that, we can use `anti_join()`, a filtering join --- ## Filtering joins Filtering joins match observations in the same way as mutating joins, but affect the observations, not the variables. There are two types: - `anti_join(x, y, by = "key")` drops all observations in `x` that have a match in `y`. - `semi_join(x, y, by = "key")` keeps all observations in `x` that have a match in `y`. --- ## `anti_join(x, y, by = "key")` .pull-left-small[ ```r anti_join(df1, df2, by="id") # no values for id in df2 ``` ``` ## id trt value ## 1 1 A 5 ## 2 2 B 3 ## 3 3 C 7 ## 4 6 C 3 ``` ```r anti_join(df2, df1, by="id") # no values for id in df1 ``` ``` ## id stress bpm ## 1 7 0 48 ## 2 7 1 110 ``` ] .pull-right-large[ - keeps rows the *don't* have a match - be careful, it is NOT symmetric! - useful for diagnosing join mismatches. ] --- class: yourturn # Your turn For this exercise, use the data from the `classdata` package - Load the `classdata` package into your R session. - Investigate data sets `box` and `budget`. - Join the two datasets to incorporate the release date of movies into the box office gross. - Check on the dimensions of the data sets before and after the join. Where are potential problems? - Use `anti_join` to detect problematic cases. --- class: inverse, middle, center # As requested, <br>more examples! --- ## `nycflights13` The `nycflights13` package contains 4 data sets related to the `flights` data that we have used previously. - `airlines` lets you look up the full carrier name from its abbreviated code - `airports` gives information about each airport, identified by the faa airport code: - `planes` gives information about each plane, identified by its `tailnum`: - `weather` gives the weather at each NYC airport for each hour: --- - `airlines` lets you look up the full carrier name from its abbreviated code ``` ## # A tibble: 2 x 2 ## carrier name ## <chr> <chr> ## 1 9E Endeavor Air Inc. ## 2 AA American Airlines Inc. ``` - `airports` gives information about each airport, identified by the faa airport code: ``` ## # A tibble: 2 x 8 ## faa name lat lon alt tz dst tzone ## <chr> <chr> <dbl> <dbl> <dbl> <dbl> <chr> <chr> ## 1 04G Lansdowne Airport 41.1 -80.6 1044 -5 A America/New_Yo… ## 2 06A Moton Field Municipal Airport 32.5 -85.7 264 -6 A America/Chicago ``` - `planes` gives information about each plane, identified by its `tailnum`: ``` ## # A tibble: 2 x 9 ## tailnum year type manufacturer model engines seats speed engine ## <chr> <int> <chr> <chr> <chr> <int> <int> <int> <chr> ## 1 N10156 2004 Fixed wing mul… EMBRAER EMB-14… 2 55 NA Turbo-… ## 2 N102UW 1998 Fixed wing mul… AIRBUS INDUSTR… A320-2… 2 182 NA Turbo-… ``` - `weather` gives the weather at each NYC airport for each hour: ``` ## # A tibble: 2 x 15 ## origin year month day hour temp dewp humid wind_dir wind_speed wind_gust ## <chr> <int> <int> <int> <int> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> ## 1 EWR 2013 1 1 1 39.0 26.1 59.4 270 10.4 NA ## 2 EWR 2013 1 1 2 39.0 27.0 61.6 250 8.06 NA ## # … with 4 more variables: precip <dbl>, pressure <dbl>, visib <dbl>, ## # time_hour <dttm> ``` --- background-image: url(https://d33wubrfki0l68.cloudfront.net/245292d1ea724f6c3fd8a92063dcd7bfb9758d02/5751b/diagrams/relational-nycflights.png) One way to show the relationships between the different tables is with a drawing: --- ## `nycflights13` relations - `flights` connects to `planes` via a single variable, `tailnum.` - `flights` connects to `airlines` through the `carrier` variable. - `flights` connects to `airports` in two ways: via the `origin` and `dest` variables. - `flights` connects to `weather` via `origin` (the location), and `year`, `month`, `day` and `hour` (the time). --- ## defining the variables to join by The default, `by = NULL`, uses all variables that appear in both tables. For example, the `flights` and `weather` tables have the common vaiables `year`, `month`, `day`, `hour`, and `origin`, but we do not need to define this when calling `left_join()`: ```r flights %>% left_join(weather) ``` ``` ## Joining, by = c("year", "month", "day", "origin", "hour", "time_hour") ``` ``` ## # A tibble: 336,776 x 28 ## year month day dep_time sched_dep_time dep_delay arr_time sched_arr_time ## <int> <int> <int> <int> <int> <dbl> <int> <int> ## 1 2013 1 1 517 515 2 830 819 ## 2 2013 1 1 533 529 4 850 830 ## 3 2013 1 1 542 540 2 923 850 ## 4 2013 1 1 544 545 -1 1004 1022 ## 5 2013 1 1 554 600 -6 812 837 ## 6 2013 1 1 554 558 -4 740 728 ## 7 2013 1 1 555 600 -5 913 854 ## 8 2013 1 1 557 600 -3 709 723 ## 9 2013 1 1 557 600 -3 838 846 ## 10 2013 1 1 558 600 -2 753 745 ## # … with 336,766 more rows, and 20 more variables: arr_delay <dbl>, carrier <chr>, ## # flight <int>, tailnum <chr>, origin <chr>, dest <chr>, air_time <dbl>, ## # distance <dbl>, hour <dbl>, minute <dbl>, time_hour <dttm>, temp <dbl>, ## # dewp <dbl>, humid <dbl>, wind_dir <dbl>, wind_speed <dbl>, wind_gust <dbl>, ## # precip <dbl>, pressure <dbl>, visib <dbl> ``` --- ## defining the variables to join by Instead, we can request that only some on the common variables be used by using a character vector, `by = "x"`: For example, `flights` and `planes` have `year` variables, but they mean different things, so we only want to join by `tailnum`: (note the changes to the `year` variables) ```r flights %>% left_join(planes, by = "tailnum") ``` ``` ## # A tibble: 336,776 x 27 ## year.x month day dep_time sched_dep_time dep_delay arr_time sched_arr_time ## <int> <int> <int> <int> <int> <dbl> <int> <int> ## 1 2013 1 1 517 515 2 830 819 ## 2 2013 1 1 533 529 4 850 830 ## 3 2013 1 1 542 540 2 923 850 ## 4 2013 1 1 544 545 -1 1004 1022 ## 5 2013 1 1 554 600 -6 812 837 ## 6 2013 1 1 554 558 -4 740 728 ## 7 2013 1 1 555 600 -5 913 854 ## 8 2013 1 1 557 600 -3 709 723 ## 9 2013 1 1 557 600 -3 838 846 ## 10 2013 1 1 558 600 -2 753 745 ## # … with 336,766 more rows, and 19 more variables: arr_delay <dbl>, carrier <chr>, ## # flight <int>, tailnum <chr>, origin <chr>, dest <chr>, air_time <dbl>, ## # distance <dbl>, hour <dbl>, minute <dbl>, time_hour <dttm>, year.y <int>, ## # type <chr>, manufacturer <chr>, model <chr>, engines <int>, seats <int>, ## # speed <int>, engine <chr> ``` --- ## defining the variables to join by We can use a named character vector, `by = c("a" = "b")`, when the key is not named the same in the two table. This will match variable `a` in table `x` to variable `b` in table `y`. The variables from `x` will be used in the output. We want to combine `flights` with `airports` which contains the location (`lat` and `lon`) of each airport, but each flight has an `origin` and `destination` airport, so we need to specify which one we want to join to: ```r flights %>% left_join(airports, by = c("dest" = "faa")) ``` ``` ## # A tibble: 336,776 x 26 ## year month day dep_time sched_dep_time dep_delay arr_time sched_arr_time ## <int> <int> <int> <int> <int> <dbl> <int> <int> ## 1 2013 1 1 517 515 2 830 819 ## 2 2013 1 1 533 529 4 850 830 ## 3 2013 1 1 542 540 2 923 850 ## 4 2013 1 1 544 545 -1 1004 1022 ## 5 2013 1 1 554 600 -6 812 837 ## 6 2013 1 1 554 558 -4 740 728 ## 7 2013 1 1 555 600 -5 913 854 ## 8 2013 1 1 557 600 -3 709 723 ## 9 2013 1 1 557 600 -3 838 846 ## 10 2013 1 1 558 600 -2 753 745 ## # … with 336,766 more rows, and 18 more variables: arr_delay <dbl>, carrier <chr>, ## # flight <int>, tailnum <chr>, origin <chr>, dest <chr>, air_time <dbl>, ## # distance <dbl>, hour <dbl>, minute <dbl>, time_hour <dttm>, name <chr>, ## # lat <dbl>, lon <dbl>, alt <dbl>, tz <dbl>, dst <chr>, tzone <chr> ``` --- class: yourturn # YOUR TURN For this exercise, use the data from the `nycflight13` package. - Add the location of the origin and destination (i.e. the `lat` and `lon`) from the `airports` data to the `flights` data - Is there a relationship between the age of a plane and its arrival delays? - Join the tables `flights` and `planes` and calculate the average arrival delay for each age of a flight. Since there are few planes older than 25 years, truncate age at 25 years. - Plot age against the average arrival delay. - What weather conditions make it more likely to see a departure delay? - Join the tables `flights` and `weather`, calculate the mean departure delay for each amount of precipitation, and plot your results. --- ## Add'l filtering join examples Recall that anti-joins are useful for diagnosing join mismatches. For example, when connecting `flights` and `planes`, you might be interested to know that there are many `flights` that don’t have a match in `planes`: ```r flights %>% anti_join(planes, by = "tailnum") %>% count(tailnum, sort = TRUE) ``` ``` ## # A tibble: 722 x 2 ## tailnum n ## <chr> <int> ## 1 <NA> 2512 ## 2 N725MQ 575 ## 3 N722MQ 513 ## 4 N723MQ 507 ## 5 N713MQ 483 ## 6 N735MQ 396 ## 7 N0EGMQ 371 ## 8 N534MQ 364 ## 9 N542MQ 363 ## 10 N531MQ 349 ## # … with 712 more rows ``` --- class: yourturn # YOUR TURN For this exercise, use the data from the `nycflight13` package. What does `anti_join(flights, airports, by = c("dest" = "faa"))` tell you? What does `anti_join(airports, flights, by = c("faa" = "dest"))` tell you? --- ## Resources - reference/document: http://dplyr.tidyverse.org/reference/ - RStudio cheat sheet for [dplyr](https://github.com/rstudio/cheatsheets/raw/master/data-transformation.pdf)